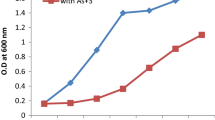
Abstract
Bacillus thuringiensis PW-05 was isolated from the Odisha coast and was found to resist 50 ppm of Hg as HgCl2 as well as higher concentrations of CdCl2, ZnSO4, PbNO3 and Na2HAsO4. Resistance towards several antibiotics, viz amoxycillin, ampicillin, methicillin, azithromycin and cephradine (CV) was also observed. The mer operon possessed by most of the mercury-resistant bacteria was also found in this isolate. Atomic absorption spectroscopy revealed that the isolate can volatilize >90 % of inorganic mercury. It showed biofilm formation in the presence of 50 ppm HgCl2 and can produce exopolysaccharide under same conditions. The isolate was found to volatilize mercury efficiently under a wide range of environmental parameters, i.e. pH (7 to 8), temperature (25 °C to 40 °C) and salinity (5 to 25 ppt). merA gene expression has been confirmed by real-time reverse transcriptase PCR study. Fourier transform infrared study revealed that –SH and –COOH groups play a major role in the process of adaptation to Hg. Hence, this isolate B. thuringiensis PW-05 shows an interesting potential for bioremediation of mercury.








Similar content being viewed by others
References
Aram M, Sharifi A, Kafeelzadeh F, Naghmachi M, Yasari E (2012) Isolating mercury-resistant bacteria from Lake Maharloo. Int J Biol 4:63–71
Bafana A, Krishnamurthi K, Patil M, Chakrabarti T (2010) Heavy metal resistance in Arthrobacter ramosus strain G2 isolated from mercuric salt-contaminated soil. J Hazard Mater 177:481–486
Barkay T, Miller SM, Summers AO (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27:355–384
Bauer A, Kirby W, Sherris J, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Path 45:493–496
Bogdanova ES, Bass IA, Minakhin LS, Petrova MA, Mindlin SZ, Volodin AA, Kalyaeva ES, Tiedjet JM, Hobman JL, Brown NL, Nikiforov VG (1998) Horizontal spread of mer operons among gram-positive bacteria in natural environments. Microbiol 144:609–620
Chikere CB, Chikere BO, Okpokwasili GC (2012) Bioreactor-based bioremediation of hydrocarbon-polluted Niger Delta marine sediment, Nigeria. 3. Biotech 2:53–66
Christensen GD, Simpson WA, Bisno AL, Beachey EH (1982) Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun 37:318–326
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175
Clinical and Laboratory Standards. Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, seventh ed., Approved Standard M7-A7, CLSI, Wayne, PA, USA, 2006.
Coral MNU, Korkmaz H, Arikan B, Coral G (2005) Plasmid mediated heavy metal resistances in Enterobacter spp. isolated from Sofulu landfill, in Adana, Turkey. Ann Microbiol 55:175–179
Das N, Vimala R, Karthika P (2008) Biosorption of heavy metals- an overview. Indian J Biotechnol 7:159–169
Dash HR, Das S (2012) Bioremediation of mercury and importance of bacterial mer genes. Int Biodeterior Biodeg 75:207–213
Dash HR, Mangwani N, Chakraborty J, Kumari S, Das S (2013) Marine bacteria: potential candidates for enhanced bioremediation. Appl Microbiol Biotechnol 97:561–571
De Souza MJ, Nair S, Loka bharathi PA, Chandramohan D (2006) Metal and antibiotic-resistance in psychrotrophic bacteria from Antarctic Marine waters. Ecotoxicol 15:379–384
De J, Ramaiah N (2007) Characterization of marine bacteria highly resistant to mercury exhibiting multiple resistances to toxic chemicals. Ecol Ind 7:511–520
De J, Ramaiah N, Mesquita A, Verlekar XN (2003) Tolerance to various toxicants by marine bacteria highly resistant to mercury. Mar Biotechnol 5:185–193
De J, Ramaiah N, Vardanyan L (2008) Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar Biotechnol 10:471–477
Deng X, Wang P (2012) Isolation of marine bacteria highly resistant to mercury and their bioaccumulation process. Bioresour Technol 121:342–347
Dietz R, Outridge PM, Hobson KA (2009) Anthropogenic contributions to mercury levels in present-day Arctic animals—a review. Sci Total Environ. doi:10.1016/j.scitotenv.2009.08.036
Dong W, Bian Y, Liang L, Gu B (2011) Binding constants of mercury and dissolved organic matter determined by a modified ion exchange technique. Environ Sci Technol 45:3576–3583
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Jain K, Parida S, Mangwani N, Dash HR, Das S (2013) Isolation and characterization of biofilm-forming bacteria and associated extracellular polymeric substances from oral cavity. Ann Microbiol DOI. doi:10.1007/s13213-013-0618-9
Jordjevic D, Wiedmann M, McLandsborough LA (2002) Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68:2950–2958
Kargar M, Jahromi MZ, Najafian M, Khajeaian P, Nahavandi R, Jahromi SR, Firoozinia M (2012) Identification and molecular analysis of mercury resistant bacteria in Kor River, Iran. African J Biotechnol 11:6710–6717
Ki JS, Zhang W, Qian PY (2009) Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J Microbiol Met 77:48–57
Krajewska B (2008) Mono- (Ag, Hg) and di- (Cu, Hg) valent metal ions effects on the activity of jack bean urease. Probing the modes of metal binding to the enzyme. J Enzyme Inhib Med Chem 23:535–542
Liebert CA, Hall RM, Summers AO (1999) Transposon Tn21, flagship of the floating genome. 63:507–522.
Mack D, Nedelmann M, Krokotsch A, Schwarkopf A, Heesemann J, Laufs R (1994) Charecterization of transposon mutants of biofilm producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intracellular adhesion. Inf Imm 62:3244–3253
Manceau A, Nagy KL (2008) Relationships between Hg(II)-S bond distance and Hg(II) coordination in thiolates. Dalton Trans 11:1421–1425
Mangwani N, Dash HR, Chauhan A, Das S (2012) Bacterial quorum sensing: functional features and potential applications in biotechnology. J Mol Microbiol Biotechnol 22:215–227
Mindlin SZ, Bass IA, Bogdanova ES, Gorlenko ZM, Kalyaeva ES, Petrova MA, Nikiforov VG (2002) Horizontal transfer of mercury resistance genes in environmental bacterial populations. Mol Biol 36:160–170
Mindlin S, Minakhin L, Petrova M, Kholodii G, Minakhina S, Gorlenko Z, Nikiforov V (2005) Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Res Microbiol 156:994–1004
Mirzaei N, Kafilzadeh F, Kargar M (2008) Isolation and identification of mercury resistant bacteria from Kor River, Iran. J Biol Sc 8:935–939
Missimer JH, Steinmetz MO, Baron R, Winkler FK, Kammerer RA, Daura X, Gunsteren WFV (2007) Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci 16:1349–1359
Mortazavi S, Rezaee A, Khavanin A, Varmazyar S, Jafarzadeh M (2005) Removal of mercuric chloride by a mercury resistant Pseudomonas putida strain. J Biol Sc 5:269–273
Naik MM, Pandey A, Dubey SK (2012) Bioremediation of metals mediated by marine bacteria. In Microorganisms in Environmental Management Ed. Satyanarayana, T., and Johri, B. N. :665–682.
Nakamura K, Nakahara H (1988) Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl Environ Microbiol 54:2871–2873
Nascimento AMA, Chartone-Souza E (2003) Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Gen Mol Res 2:92–101
Naz N, Young HK, Ahmed N, Gadd GM (2005) Cadmium accumulation and DNA homology with metal resistance genes in sulfate-reducing bacteria. Appl Environ Microbiol 71:4610–4618
Nazaret S, Jeffrey WH, Saouter E, Von Haven R, Barkay T (1994) merA gene expression in aquatic environments measured by mRNA production and Hg (II) volatilization. Appl Environ Microbiol 60(11): 4059–4065
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Ann Rev Microbiol 54:49–79
Ogungbenle HN, Oshodi AA, Oladimeji MO (2009) The proximate and effect of salt applications on some functional properties of quinoa (Chenopodium quinoa) flour. Pakistan J Nutr 8:49–52
Osborn AM, Bruce KD, Strike P, Ritchie DA (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev 19:239–262
Pahan K, Ghosh DK, Chaudhuri J, Gachhui R, Ray S, Mandal A (1995) Mercury detoxifying enzymes within endospores of a broad-spectrum mercury resistant Bacillus pasteurii strain DR2. J Biosc 20:83–88
Parry E (2006) A broad-spectrum mer operon in a multi-drug resistant strain of the fish pathogen, Aeromonas salmonicida. Undergraduate Research Symposium. Paper 52. http://digitalcommons.colby.edu/ugrs/52.
Partridge SR, Brown HJ, Stokes HW (2001) Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother 45:1263–1270
Pike R, Lucas V, Stapleton P, Gilthorpe MS, Roberts G, Rowbury R, Richards H, Mullany P, Wilson M (2002) Prevalence and antibiotic resistance profile of mercury-resistant oral bacteria from children with or without mercury amalgam fillings. J Antimicrob Chemother 49:777–783
Poulain AJ, Chadhain SMN, Ariya PA, Amyot M, Garcia E, Campbell PGC, Zylstra GJ, Barkay T (2007) Potential for mercury reduction by microbes in the high arctic. Appl Environ Microbiol 73:2230–2238
Raphael EC, Augustina OC, Frank EO (2011) Trace metals distribution in fish tissues, bottom sediments and water from Okumeshi River in Delta State, Nigeria. Environ Res J 5:6–10
Rhykerd RL, Weaver RW, McInnes KJ (1995) Influence of salinity on bioremediation of oil in soil. Environ Poll 90:127–130
Rochelle PA, Wetherbee MK, Olson BH (1991) Distribution of DNA sequences encoding narrow and broad spectrum mercury resistance. Appl Environ Microbiol 57:1581–1589
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mo Biol Evol 4:406–425
Sambrook J, Russel DW (2001). Molecular cloning: a laboratory manual 3rd Ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY
Saurav K, Kannabiran K (2011) Biosorption of Cr(III) and Cr(VI) by Streptomyces VITSVK9 spp. Ann Microbiol 61:833–841
Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P (2004) Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol 186:427–437
Singh S, Kang SH, Mulchandani A, Chen W (2008) Bioremediation: environmental clean-up through pathway engineering. Curr Opin Biotechnol 19:437–444
Sotero-Martins A, Jesus MS, Lacerda M, Moreira JC, Filgueiras ALL, Barrocas PRG (2008) A conservative region of the mercuric reductase gene (merA) as a molecular marker of bacterial mercury resistance. Brazilian J Microbiol 39:307–310
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tribelli PM, Martino CD, Lopez NI, Iustman LJR (2012) Biofilm lifestyle enhances diesel bioremediation and biosurfactant production in the Antarctic polyhydroxyalkanoate producer Pseudomonas extremaustralis. Biodeg 23:645–651
Vu B, Chen M, Crawford RJ, Ivanova EP (2009) Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14:2535–2554
Wireman J, Liebert CA, Smith T, Summers AO (1997) Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl Environ Microbiol 63:4494–4503
Xiao-xi Z, Jian-xin T, Pei J, Hong-wei L, Zhi-min D, Xue-duan L (2010) Isolation, characterization and extraction of mer gene of Hg2+ resisting strain D2. Trans Nonferrous Met Soc China 20:507–512
Zeyaullah M, Islam B, Ali A (2010) Isolation, identification and PCR amplification of merA gene from highly polluted Yamuna river. African J Biotechnol 9:3510–3514
Zhang W, Chen L, Liu D (2012) Characterization of a marine-isolated mercury resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl Microbiol Biotechnol 93:1305–1314
Acknowledgements
Authors would like to acknowledge the authorities of NIT, Rourkela, for providing the facilities. H.R.D. and N.M. gratefully acknowledge the receipt of research fellowship from the Ministry of Human Resource Development, Government of India. S.D. thanks the Department of Biotechnology, Government of India, for the research grants on marine bacterial biofilm-based bioremediation of PAHs and heavy metals.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Robert Duran
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 48 kb)
Rights and permissions
About this article
Cite this article
Dash, H.R., Mangwani, N. & Das, S. Characterization and potential application in mercury bioremediation of highly mercury-resistant marine bacterium Bacillus thuringiensis PW-05. Environ Sci Pollut Res 21, 2642–2653 (2014). https://doi.org/10.1007/s11356-013-2206-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-013-2206-8